Computational Simulation of Photochemical Reactions in DNA
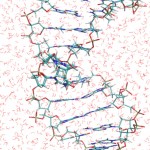
The figure shows a fragment of DNA, with the thymine dimer in the middle, sorrounded by the water molecules of the solvent.
Article: published in The Journal of Physical Chemistry Letters by Jesús I. Mendieta-Moreno and José Ortega, IFIMAC researchers and members of the Department of Theoretical Condensed Matter Physics.
The photostability of DNA is a key property for life. It is well-known that the absorption of ultraviolet (UV) radiation may result in harmful genetic lesions that affect DNA replication and transcription, ultimately causing mutations, cancer, and/or cell death. Luckily for us, cellular DNA presents remarkable stability against this photodamage: the huge majority of the absorbed photons are transformed into heat, which is transferred to the solvent without causing any lesion.
The most frequent DNA photolesion produced by sunlight is the cyclobutane thymine dimer (CTD) that is characterized by the formation of two covalent bonds between adjacent thymine bases (see Figure). In a recent collaboration, led by an IFIMAC group and published in the The Journal of Physical Chemistry Letters, this photochemical reaction has been simulated at the atomic level. The results reveal how the structure and dynamics of the DNA double-helix drastically reduce the probability of photolesion, thus protecting the integrity of the genetic code. The results also highlight the importance of properly taken into account the biomolecular environment for the study of photochemical reactions in biomolecules.
Quantum Mechanics and Molecular Mechanics
Theoretical analysis of photochemical reactions in biomolecules is a very challenging problem that requires mixing different theoretical and computational strategies. In this work, a hybrid quantum mechanics / molecular mechanics (QM/MM) method, recently developed by the authors, was used to explore the conformational and dynamical properties of the system. This method presents a very good balance between accuracy and computational efficiency, a very important property to study complex biomolecular systems. Moreover, non-adiabatic QM/MM molecular dynamics simulations were performed to study the dynamics of photo-excited DNA and analyze the atomic mechanisms of the reaction. [Full article]
Also read on UAM Gazette.