Solving the Mystery of the Strikingly Different Mechanical Response of Nucleic Acids
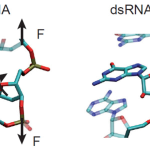
Representation of two base-pair steps to highlight the different orientation of the sugar with respect to the phosphate backbone of dsDNA (Left) and dsRNA (Right). The extra hydroxyl group in the ribose – the dsRNA sugar – changes its stereochemistry and is ultimately responsible for the opposite twist-stretch coupling.
Article: published in Proceedings of the National Academy of Sciences by J. G. Vilhena, and Ruben Perez, IFIMAC researchers and members of the Department of Theoretical Condensed Matter Physics.
The mechanical properties of nucleic acids (NAs) regulate multiple biological processes ranging from complex chromosome packing to replication of a plasmid. In vivo, NAs are not typically found in their relaxed forms. Instead, in the biological processes in which they take part, proteins wrap, bend, stretch, and twist double-stranded DNA (dsDNA) and double-stranded RNA (dsRNA) molecules. In spite of sharing a common double helix structure, single-molecule experiments have reported puzzling differences between their mechanical properties. dsRNA has a threefold softer stretching constant. More strikingly, dsDNA overwinds when stretched while dsRNA displays the expected unwinding response.
Understanding how a force induces changes in the structure of NAs at the atomic level is a challenge. In a recent publication in the Proceedings of the National Academy of Sciences (PNAS), researchers from the National Center for Biotechnology (CNB-CSIC) and from IFIMAC have used all-atom, microsecond-long molecular dynamics (MD) simulations to unveil the atomic-scale origin of the marked difference in the stretching response of dsRNA and dsDNA, their opposite twist-stretch coupling, and its nontrivial force dependence. They have implemented a new constant-force protocol, that closely mimics the single-molecule experiments, in order to simulate the structure of dsDNA and dsRNA subjected to stretching forces up to 20 pN. This methodology allows a direct determination of all of the elastic constants through the response of the average elongation, the average twist, and the coupling of their fluctuations to the applied force.
A hierarchical analysis of these simulations sheds light into the physical mechanisms that control the mechanical response. The lower dsRNA stretching resistance is linked to its more open structure, whereas the opposite twist-stretch coupling of both molecules is due to the very different evolution of the molecules’ interstrand distance with the stretching force. A reduction of this distance leads to overwinding in dsDNA. In contrast, dsRNA is not able to reduce its interstrand distance and can only elongate by unwinding. The analysis of the parameters that characterize locally the double helix shows a direct correlation between the interstrand distance and the slide, the displacement of two consecutive base pairs in the plane perpendicular to the helical axis. In turn, the different behavior of the slide parameter of dsDNA and dsRNA can be traced down to the most fundamental difference between these two molecules: the extra hydroxyl group in the dsRNA sugar that connects the nitrogenous bases to the phosphate backbone. The comprehensive atomic-scale understanding of the mechanical response of NAs achieved in this study highlights MD simulations as a powerful tool to unveil the connection between forces and structure of NAs and, possibly, to gain insight into the associated changes in their biological functionality. [Full article]
References
- Alberto Marin-Gonzalez, Jose Guilherme Vilhena, Rubén Pérez, and Fernando Moreno-Herrero. Understanding the mechanical response of double-stranded DNA and RNA under constant stretching forces using all-atom molecular dynamics. PNAS, (2017).