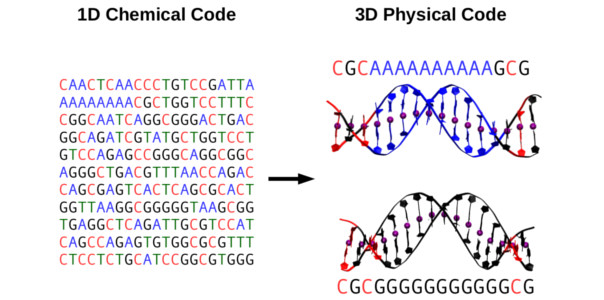Towards Deciphering the Physical Code of DNA
Article: published in Physical Review Letters by Rubén Pérez, IFIMAC researcher and member of the Department of Theoretical Condensed Matter Physics.
The genomic material needs to be precisely organized and access to its information carefully regulated in order to ensure proper functioning of a cell. These instructions are encoded in the DNA, although not in the form of the well-known three-letter words (or codons) that chemically code for proteins. In this work we found a striking connection between DNA sequence, structure and flexibility: the sequence can specifically modify the flexibility by means of concerted changes in the structure of the double-helix. Our state-of-the-art computer simulations showed that DNA molecules with different sequences substantially differed in their extension. This was a consequence of an internal curvature of the DNA that we denoted crookedness. As we increased the force, sequences that were initially more compressed – or more crooked – were able to elongate by a greater amount that those that were already elongated at low forces. Crookedness works as a reservoir of elastic energy that is directly encoded in the nucleotide sequence. Remarkably, sequences that lacked this reservoir had been previously described to destabilize nucleosomes, the first step of DNA compaction into chromosomes. As a consequence, these DNA regions are known to be highly accessible to the cellular machinery, triggering many key biological processes. [Full article]
Research Highlight in Nature Reviews Physics: DNA’s crooked path.